Publications
Genetic heterogeneity and co-occurring driver mutations impact clinical outcomes in blood cancers, but predicting the emergent effect of co-occurring mutations that impact multiple complex and interacting signalling networks is challenging. Here, we used mathematical models to predict the impact of co-occurring mutations on cellular signalling and cell fates in diffuse large B cell lymphoma and multiple myeloma. Simulations predicted adverse impact on clinical prognosis when combinations of mutations induced both anti-apoptotic (AA) and pro-proliferative (PP) signalling. We integrated patient-specific mutational profiles into personalised lymphoma models, and identified patients characterised by simultaneous upregulation of anti-apoptotic and pro-proliferative (AAPP) signalling in all genomic and cell-of-origin classifications (8-25% of patients). In a discovery cohort and two validation cohorts, patients with upregulation of neither, one (AA or PP), or both (AAPP) signalling states had good, intermediate and poor prognosis respectively. Combining AAPP signalling with genetic or clinical prognostic predictors reliably stratified patients into striking prognostic categories. AAPP patients in poor prognosis genetic clusters had 7.8 months median overall survival, while patients lacking both features had 90% overall survival at 120 months in a validation cohort. Personalised computational models enable identification of novel risk-stratified patient subgroups, providing a valuable tool for future risk-adapted clinical trials.
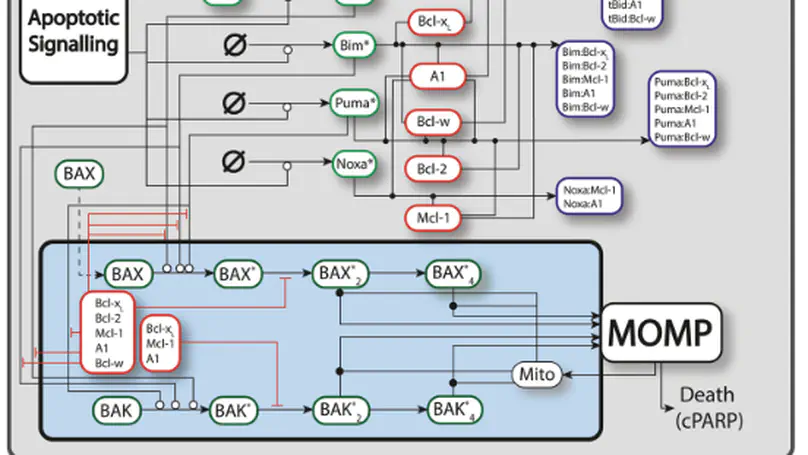
In healthy cells, pro- and anti-apoptotic BCL2 family and BH3-only proteins are expressed in a delicate equilibrium. In contrast, this homeostasis is frequently perturbed in cancer cells due to the overexpression of anti-apoptotic BCL2 family proteins. Variability in the expression and sequestration of these proteins in Diffuse Large B cell Lymphoma (DLBCL) likely contributes to variability in response to BH3-mimetics. Successful deployment of BH3-mimetics in DLBCL requires reliable predictions of which lymphoma cells will respond. Here we show that a computational systems biology approach enables accurate prediction of the sensitivity of DLBCL cells to BH3-mimetics. We found that fractional killing of DLBCL, can be explained by cell-to-cell variability in the molecular abundances of signaling proteins. Importantly, by combining protein interaction data with a knowledge of genetic lesions in DLBCL cells, our in silico models accurately predict in vitro response to BH3-mimetics. Furthermore, through virtual DLBCL cells we predict synergistic combinations of BH3-mimetics, which we then experimentally validated. These results show that computational systems biology models of apoptotic signaling, when constrained by experimental data, can facilitate the rational assignment of efficacious targeted inhibitors in B cell malignancies, paving the way for development of more personalized approaches to treatment.
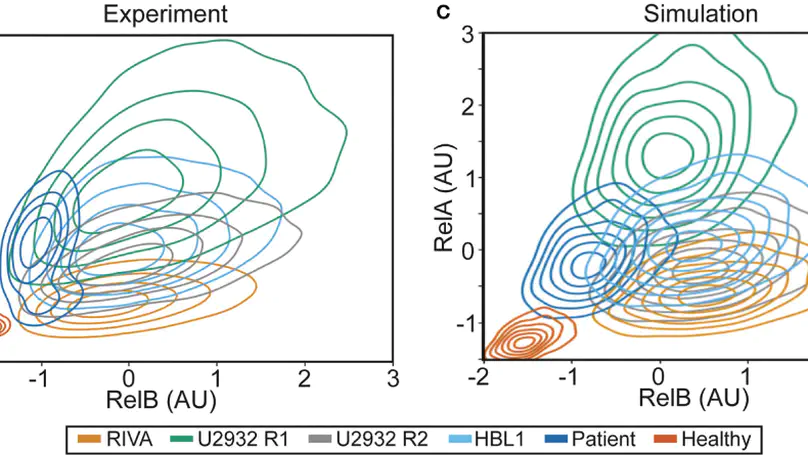
Introduction. Improving treatments for Diffuse Large B-Cell Lymphoma (DLBCL) is challenged by the vast heterogeneity of the disease. Nuclear factor-κB (NF-κB) is frequently aberrantly activated in DLBCL. Transcriptionally active NF-κB is a dimer containing either RelA, RelB or cRel, but the variability in the composition of NF-κB between and within DLBCL cell populations is not known. Results. Here we describe a new flow cytometry-based analysis technique termed “NF-κB fingerprinting” and demonstrate its applicability to DLBCL cell lines, DLBCL core-needle biopsy samples, and healthy donor blood samples. We find each of these cell populations has a unique NF-κB fingerprint and that widely used cell-of-origin classifications are inadequate to capture NF-κB heterogeneity in DLBCL. Computational modeling predicts that RelA is a key determinant of response to microenvironmental stimuli, and we experimentally identify substantial variability in RelA between and within ABC-DLBCL cell lines. We find that when we incorporate NF-κB fingerprints and mutational information into computational models we can predict how heterogeneous DLBCL cell populations respond to microenvironmental stimuli, and we validate these predictions experimentally. Discussion. Our results show that the composition of NF-κB is highly heterogeneous in DLBCL and predictive of how DLBCL cells will respond to microenvironmental stimuli. We find that commonly occurring mutations in the NF-κB signaling pathway reduce DLBCL’s response to microenvironmental stimuli. NF-κB fingerprinting is a widely applicable analysis technique to quantify NF-κB heterogeneity in B cell malignancies that reveals functionally significant differences in NF-κB composition within and between cell populations.
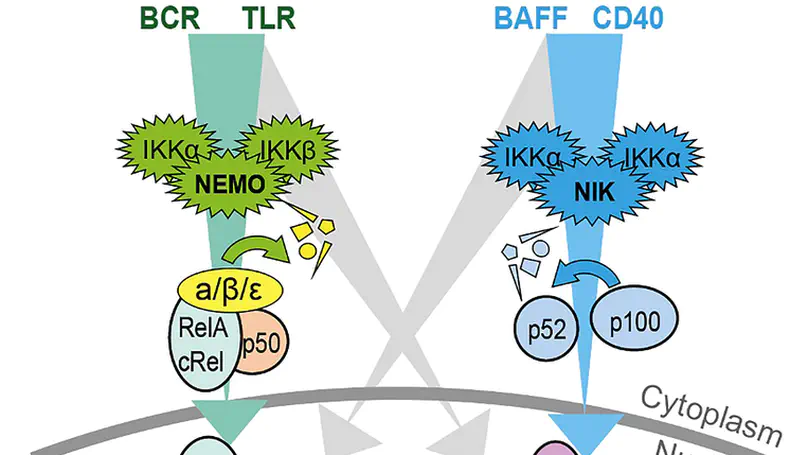
Chronic lymphocytic leukemia (CLL) is the most prevalent type of leukemia in the western world. Despite the positive clinical effects of new targeted therapies, CLL still remains an incurable and refractory disease and resistance to treatments are commonly encountered. The Nuclear Factor-Kappa B (NF-κB) transcription factor has been implicated in the pathology of CLL, with high levels of NF-κB associated with disease progression and drug resistance. This aberrant NF-κB activation can be caused by genetic mutations in the tumor cells and microenvironmental factors, which promote NF-κB signaling. Activation can be induced via two distinct pathways, the canonical and non-canonical pathway, which result in tumor cell proliferation, survival and drug resistance. Therefore, understanding how the CLL microenvironment drives NF-κB activation is important for deciphering how CLL cells evade treatment and may aid the development of novel targeting therapeutics. The CLL microenvironment is comprised of various cells, including nurse like cells, mesenchymal stromal cells, follicular dendritic cells and CD4+ T cells. By activating different receptors, including the B cell receptor and CD40, these cells cause overactivity of the canonical and non-canonical NF-κB pathways. Within this review, we will explore the different components of the CLL microenvironment that drive the NF-κB pathway, investigating how this knowledge is being translated in the development of new therapeutics.
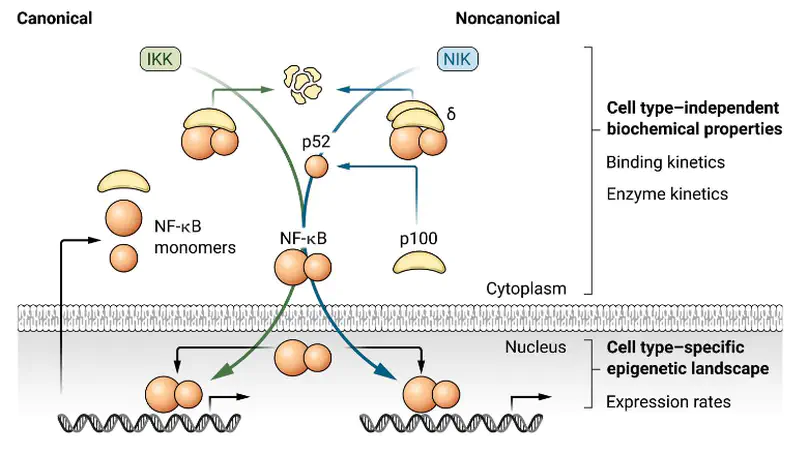
The nuclear factor κB (NF-κB) system is critical for various biological functions in numerous cell types, including the inflammatory response, cell proliferation, survival, differentiation, and pathogenic responses. Each cell type is characterized by a subset of 15 NF-κB dimers whose activity is regulated in a stimulus-responsive manner. Numerous studies have produced different mathematical models that account for cell type–specific NF-κB activities. However, whereas the concentrations or abundances of NF-κB subunits may differ between cell types, the biochemical interactions that constitute the NF-κB signaling system do not. Here, we synthesized a consensus mathematical model of the NF-κB multidimer system, which could account for the cell type–specific repertoires of NF-κB dimers and their cell type–specific activation and cross-talk. Our review demonstrates that these distinct cell type–specific properties of NF-κB signaling can be explained largely as emergent effects of the cell type–specific expression of NF-κB monomers. The consensus systems model represents a knowledge base that may be used to gain insights into the control and function of NF-κB in diverse physiological and pathological scenarios and that describes a path for generating similar regulatory knowledge bases for other pleiotropic signaling systems.
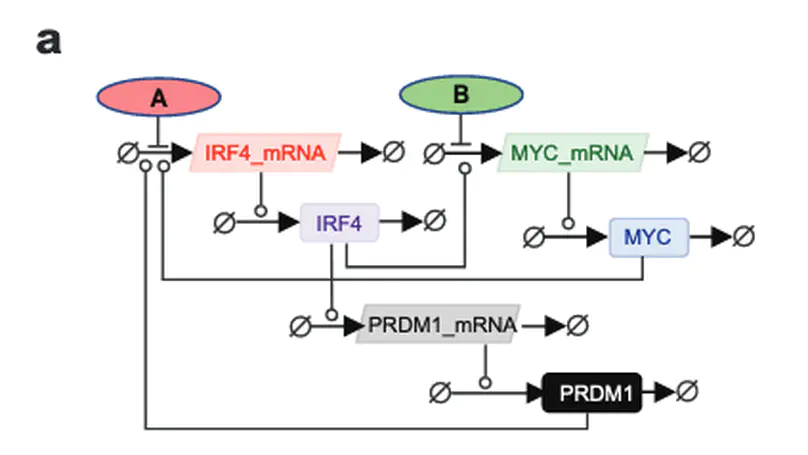
B-cell progenitor fate determinant interferon regulatory factor 4 (IRF4) exerts key roles in the pathogenesis and progression of multiple myeloma (MM), a currently incurable plasma cell malignancy. Aberrant expression of IRF4 and the establishment of a positive auto-regulatory loop with oncogene MYC, drives a MM specific gene-expression programme leading to the abnormal expansion of malignant immature plasma cells. Targeting the IRF4-MYC oncogenic loop has the potential to provide a selective and effective therapy for MM. Here we evaluate the use of bromodomain inhibitors to target the IRF4-MYC axis through combined inhibition of their known epigenetic regulators, BRD4 and CBP/EP300. Although all inhibitors induced cell death, we found no synergistic effect of targeting both of these regulators on the viability of MM cell-lines. Importantly, for all inhibitors over a time period up to 72 hours, we detected reduced IRF4 mRNA, but a limited decrease in IRF4 protein expression or mRNA levels of downstream target genes. This indicates that inhibitor-induced loss of cell viability is not mediated through reduced IRF4 protein expression, as previously proposed. Further analysis revealed a long half-life of IRF4 protein in MM cells. In support of our experimental observations, gene network modelling of MM suggests that bromodomain inhibition is exerted primarily through MYC and not IRF4. These findings suggest that despite the autofeedback positive regulatory loop between IRF4 and MYC, bromodomain inhibitors are not effective at targeting IRF4 in MM and that novel therapeutic strategies should focus on the direct inhibition or degradation of IRF4.