The NF-κB multidimer system model: A knowledge base to explore diverse biological contexts
Abstract
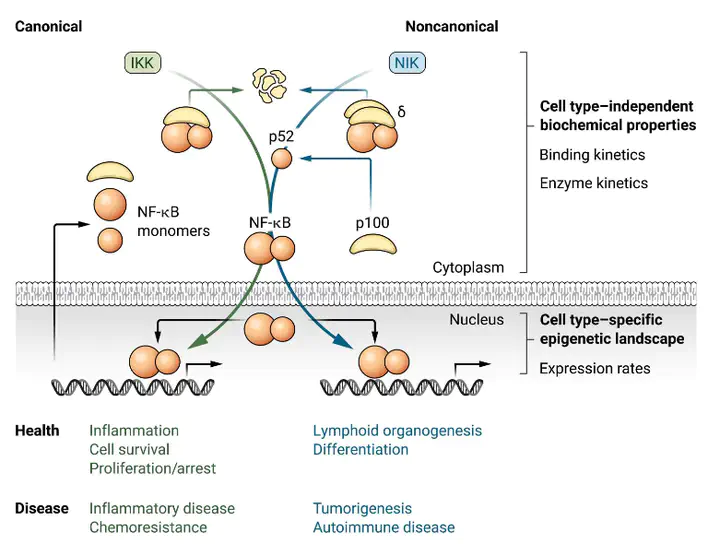
The nuclear factor κB (NF-κB) system is critical for various biological functions in numerous cell types, including the inflammatory response, cell proliferation, survival, differentiation, and pathogenic responses. Each cell type is characterized by a subset of 15 NF-κB dimers whose activity is regulated in a stimulus-responsive manner. Numerous studies have produced different mathematical models that account for cell type–specific NF-κB activities. However, whereas the concentrations or abundances of NF-κB subunits may differ between cell types, the biochemical interactions that constitute the NF-κB signaling system do not. Here, we synthesized a consensus mathematical model of the NF-κB multidimer system, which could account for the cell type–specific repertoires of NF-κB dimers and their cell type–specific activation and cross-talk. Our review demonstrates that these distinct cell type–specific properties of NF-κB signaling can be explained largely as emergent effects of the cell type–specific expression of NF-κB monomers. The consensus systems model represents a knowledge base that may be used to gain insights into the control and function of NF-κB in diverse physiological and pathological scenarios and that describes a path for generating similar regulatory knowledge bases for other pleiotropic signaling systems.
We used a computational model of NF-kB as a knowledge base, to review how a highly conserved system can do very different things in different contexts. Different cell types have different NF-kB states and responses to stimuli… why?
- The interactions are the same,
- The fundamental kinetic rates are probably the same,
- But epigenetics will lead to different expression rates and therefore abundances of NF-kB monomers.
Is cell-type specific expression of NF-kB monomers enough to explain the vast and varied data out there?
We used a model to figure this out.
We incorporated cell type-specific gene expression data into the model and review the literature, comparing virtual cells to what is known. We reviewed the literature on NF-kB and its response to canonical and non-canonical activating stimuli in different cell types.
We found that a single conserved signalling network can capture this behavior, by adjusting only cell type–specific expression of NF-κB monomers. Finally, we turned to cancer, and mutations that occur in cancers including B cell lymphomas. We found that the single conserved model can also capture what is known about how mutations derail the system. We just needed to adjust the parameter affected by the mutation.